Improving the performance of DL_MONTE for large-scale simulations
ARCHER2-eCSE01-19
PI: Prof Steve Parker (University of Bath)
Co-I(s): Dr John A Purton (Science and Technology Facilities Council), Prof Tina Duren (University of Bath)
Technical staff: Dr Tom L Underwood (University of Bath)
Subject Area:
This image is a snapshot from a simulation of a paracetamol crystal surface in contact with water, at conditions where bubbles form. This simulation was performed as part of an investigation into surfaces of crystals of pharmaceutical molecules. The simulation was performed on ARCHER 2 using DL_MONTE. (Courtesy of Jay Zhou, University of Bath)
Monte Carlo molecular simulation (MCMS) entails using random numbers to calculate properties of solids or fluids at the atomic scale. A key aspect of MCMS is that it evolves the positions of atoms through time in an unrealistic manner. For instance, during a simulation the positions of atoms belonging to different elements could be swapped, or molecules could be inserted into, or deleted from, the system. In certain situations, this aspect of MCMS gives it an advantage over ‘standard’ methods in which atoms are moved in a realistic manner. For this reason, MCMS is the method of choice for studying various physical phenomena of key relevance to technology; common applications of MCMS include quantifying the amount of a gas which adsorbs to a surface or material, and calculating the atomic-scale properties of fluids and mixtures.
Modern high-performance computing (HPC) facilities such as ARCHER 2 have the potential to enable MCMS to be used to calculate atomic-scale properties to a higher accuracy than was previously possible. However, this prospect requires MCMS software which can effectively make use of the computing power of large HPC facilities. Unfortunately, a current limitation of most existing MCMS programs is that they are not geared towards this.
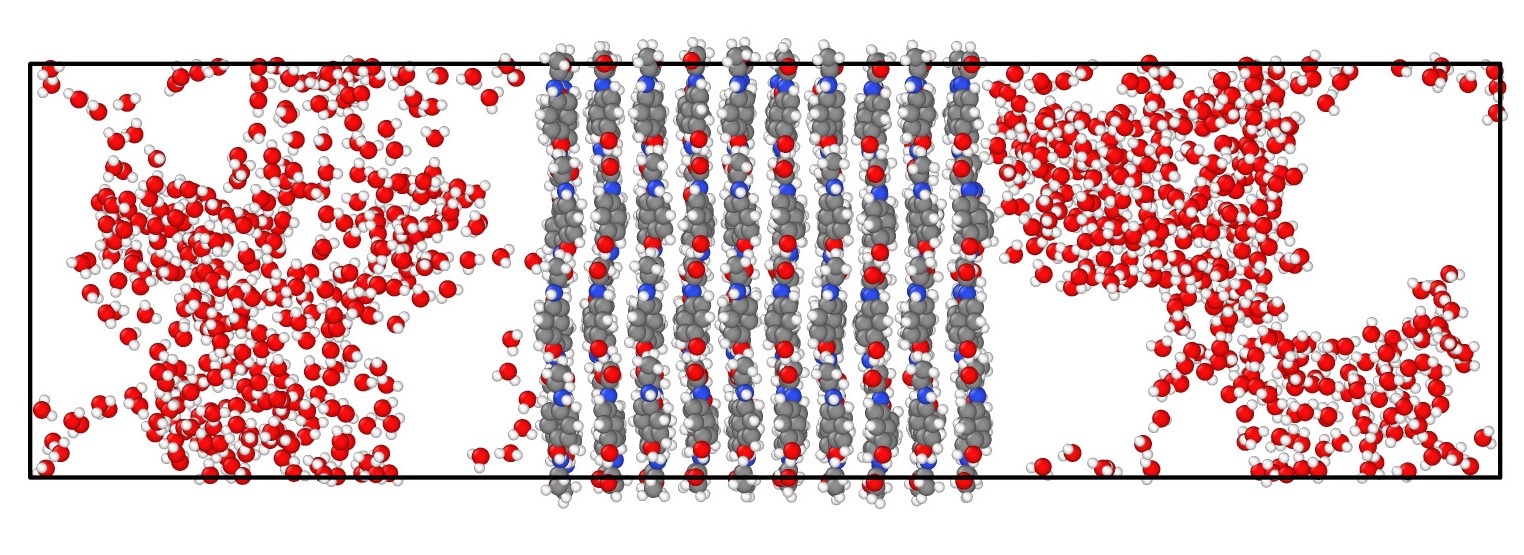
The aim of this project was to address this. We worked on the open-source MCMS computer program DL_MONTE, improving its capabilities with regards to ‘large-scale’ calculations afforded by the latest HPC facilities. Firstly, we implemented new algorithms within DL_MONTE which enable it to efficiently simulate systems containing very large numbers of atoms. Secondly, we implemented a new method in DL_MONTE which enables it to apply key capabilities to larger, more complex molecules. Finally, we greatly improved DL_MONTE’s ability to use multiple computer cores in parallel to perform calculations more quickly. These improvements will enable the scientific community to use MCMS in conjunction with facilities such as ARCHER 2 to perform more accurate studies regarding, e.g. adsorption at surfaces, with broad scientific benefits in the long term.
Information about the code
DL_MONTE is open source, and is hosted on GitLab, where documentation and tutorials pertaining to DL_MONTE, as well as its source code (for various versions) can be found.
The latest development version of the program can be found on gitlab.
To install the program on ARCHER2, execute the following commands within the directory containing the source code:
./build: SRL dir archer2
./build: PRL dir archer
These commands compile both the serial and parallel executables of DL_MONTE, DLMONTE-SRL.X and DLMONTE-PRL.X, respectively, and house them within the bin subdirectory within the source code’s directory.