Optimising NEMO-ERSEM for High Resolution
ARCHER2-eCSE04-06
PI: Dr Jerry J C Blackford (Plymouth Marine Laboratory)
Co-I(s): Dr James Harle (National Oceanography Centre), Dr Yuri Artioli (Plymouth Marine Laboratory), Dr Susan Kay (Plymouth Marine Laboratory), Dr Gennadi Lessin (Plymouth Marine Laboratory)
Technical staff: Dr Dale Partridge (Plymouth Marine Laboratory), Mr Michael Wathen (Plymouth Marine Laboratory), Dr Andrew G Sunderland (STFC), Dr Andrew R Porter (STFC)
Subject Area:
3D coupled marine models such as NEMO-FABM-ERSEM represent the marine environment with a discrete grid of layered, (usually squared) prisms. A key characteristic of these models is their resolution, expressed as the distance between prisms and/or layers, with higher resolution generally providing more skilful simulations and better addressing issues at the scale of biological processes and stakeholder interaction. However, increasing resolution greatly impacts the computational and storage requirements for a given simulation. This introduces a balance between the desire to use the highest resolution/skill possible and the computational aspects; memory, I/O, and communication. In the case of NEMO-FABM-ERSEM, simulations can be computationally expensive, and by dedicating time to ensure the code is as optimized as possible to run on ARCHER2 significant time and resources can be saved.
A key simulation used by multiple UK institutions covers the Northwest European Shelf. The latest generation grid for this region is the AMM15 grid, which increases the resolution from 7km to 1.5km. Using this as our benchmark we performed a series of profiling experiments to test the suitability of the code and identify any weak spots for improvement.
As you can see in Figure 1, parallel scaling is almost ideal up to 8400 cores, then begins to deteriorate due to an increase in the relative cost of data exchange between tasks. The strong scaling performance indicates that the code is generally already well optimised, and gains will need to be found elsewhere.
We found that by changing the environment in which the runs take place; through enabling striping across Lustre storage and by using collective calls for parallel I/O performance could be improved. Enabling these options led to a runtime reduction of up to 4%. Additional runtime gains could also be found by under-populating nodes, although this option comes at a greater overall cost in node hours.
These benefits are not insignificant, albeit much smaller than envisioned at the onset of this project. In addition to the above we also undertook an extensive analysis of how the code responds to changes in the number of tracer variables, reflecting an increase/decrease in the biogeochemical model complexity. Here we observed that some routines did not scale well as tracers increased, something that needs to be considering when developing the next generation of biogeochemical models.
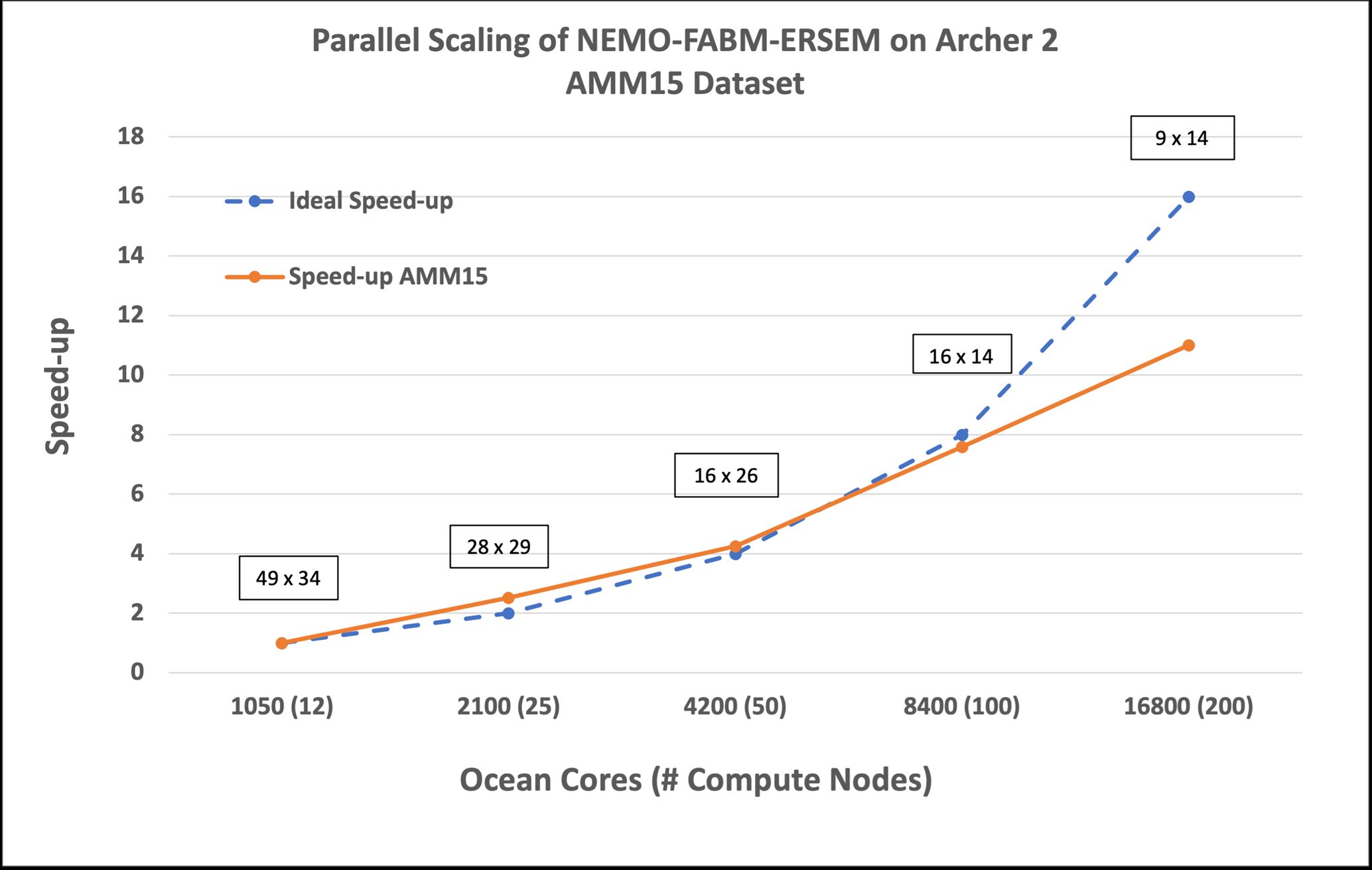
Figure 1: Parallel Scalability of NEMO-FABM-ERSEM on Archer 2 with the AMM15 Dataset.
Boxed labels represent the number of points per subdomain
Information about the code
NEMO-FABM-ERSEM is a coupled numerical modelling system for simulating the marine environment, used by many research groups and led by Plymouth Marine Laboratory, the National Oceanographic Centre and the Met Office. It couples the following modelling frameworks:
- NEMO (Nucleus for European Modelling of the Ocean), a state-of-the-art modelling framework for ocean and climate modelling
- FABM (Framework for Aquatic Biogeochemical Models), a programming framework for biogeochemical models of marine and freshwater systems
- ERSEM (European Regional Seas Ecosystem Model), a complex marine ecosystem model which addresses biogeochemical and ecological systems in many applications in global regional seas and the global ocean
This eCSE project specifically focused on the biogeochemistry component of the coupled model. Full details of how to get, compile and run the code are included in the technical report.
The executable for the project is ‘nemo’, or ‘nemo.exe’. However, this is also the case for the non-coupled version of the code. A simple search of the executable names will not distinguish between the two.